The Nobel Prize in the natural sciences often evokes a sense of a distant, unattainable world, seemingly disconnected from everyday life. However, this year’s Nobel Prize in Chemistry directly influences the work of researchers at Latvian Institute of Organic Synthesis in developing new drug compounds.
The protein structure prediction programme AlphaFold, created by Nobel laureates Demis Hassabis and John Michael Jumper from Google DeepMind, is set to significantly reduce the time required to develop new drugs. Previously, this process would typically take around 10 years, but thanks to these new methods, it could now be shortened by several years.
Dr Raitis Bobrovs, head of the Structural Biology and Drug Design Laboratory at LIOS, is one of the few researchers in Latvia who works daily with protein modelling, including using AlphaFold, developed by the Nobel laureates. For several years, he has focused on computational modelling in chemistry, recognising its enormous potential and also the opportunity to pursue his interest in mathematical and computational processes in science. In his daily work, he uses computer calculations to identify new drug molecules and optimise known ones, contributing to the earliest stages of drug development.
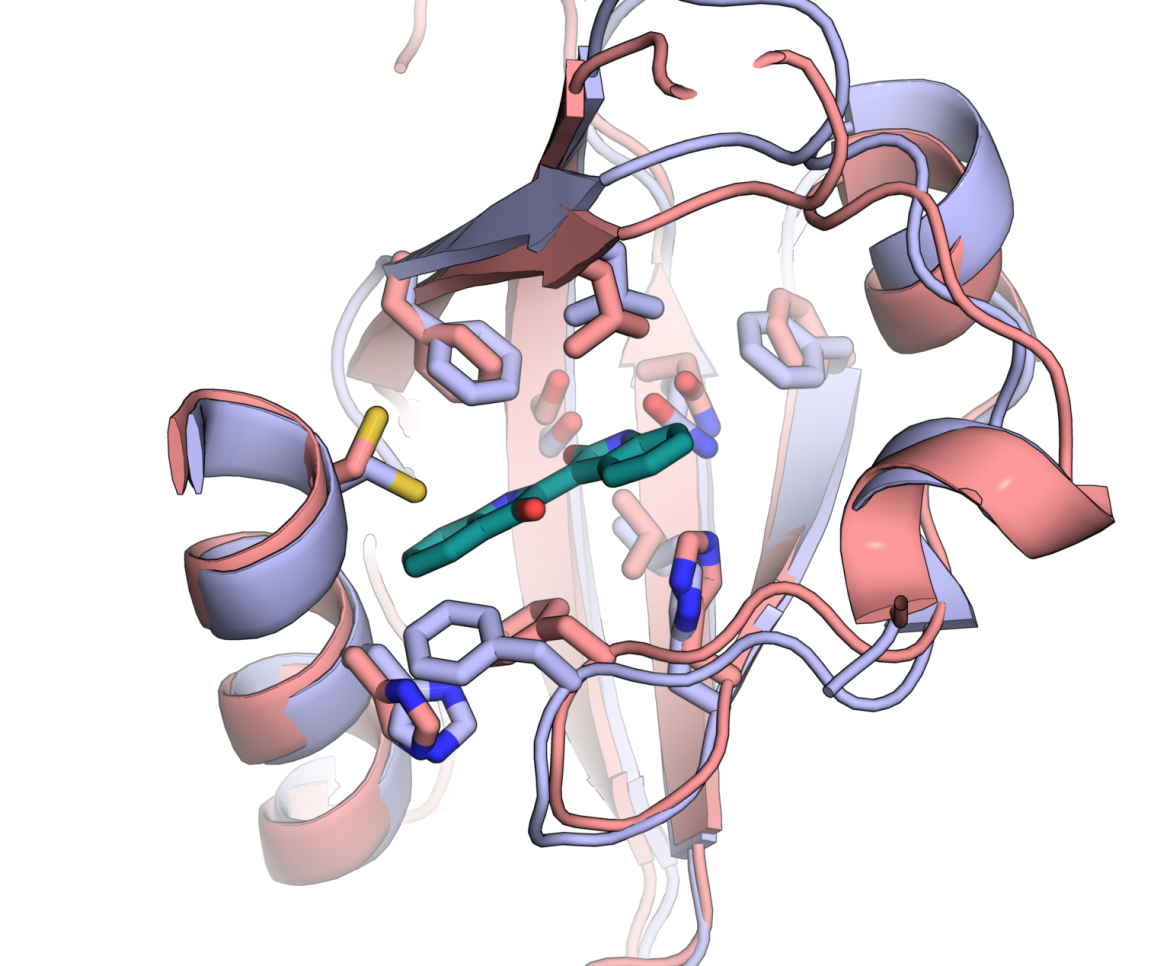
Image caption: The structure of an aryl hydrocarbon receptor (in purple), modelled by AlphaFold, which Raitis Bobrovs and his colleagues used in the discovery and development of new potential drug compounds. For comparison, the experimentally determined (and real-life confirmed) structure (in red), obtained two years later, is very similar to the one generated by computer modelling.
What do drugs target?
Drug compounds are most often designed to target specific proteins in the human body or in the pathogens that cause disease. “For example, we are currently working on developing new anti-malarial drug compounds at the institute,” explains Raitis Bobrovs, referring to one of his projects. “Biologists study the various proteins of the malaria pathogen, seeking those vital to its life functions. We, as chemists, search for ways to destroy these proteins by blocking them. To achieve this, we need to find molecules that will bind to the target protein in a specific way and disrupt its function. It is equally important to ensure that our active compounds affect only the specific pathogen’s proteins and not similar proteins in the human body.”
Unfolding and folding proteins
All living organisms contain proteins. Proteins are constantly interacting, carrying out essential processes to sustain life, but also contributing to ageing or triggering disease processes. Each protein is composed of amino acids (organic compounds). All proteins in the world are made up of the same 20 amino acids, but arranged in different sequences, resulting in a vast array of proteins with varying structures and functions. The number of proteins on this planet exceeds 200 million, each with a unique structure and shape. In 1972, American scientist Christian Anfinsen received the Nobel Prize in Chemistry for discovering that after deliberately “unfolding” protein structures, they would reassemble into precisely the same shape they originally had. He concluded that a protein’s spatial arrangement is strictly determined by its unique amino acid sequence.
From years to 2 hours
For decades, scientists struggled to find a way to observe individual proteins in nature in order to understand how to target them. Until recently, proteins were studied using X-rays, where the tiny structures were illuminated by X-ray beams and examined over a long period of time. This method of determining protein structures was highly time-consuming, meaning the early stages of drug development—when a suitable active compound is sought for a specific protein—could take several years.
This year’s Nobel laureates combined structural biology with artificial intelligence and, in 2021, created a computer programme capable of modelling and spatially visualising any protein on the planet. The two scientists input data on the structures of 200,000 proteins that had been determined so far, using this information to train the AI to identify common patterns in amino acid sequences, allowing it to predict protein structures with near-perfect accuracy and visualise them in 3D.
“This protein structure database has greatly simplified, sped up, and made more focused the work of scientists in the early stages of drug development,” says Raitis Bobrovs. “Previously, determining a protein’s structure could take months or even years. Now, using the AlphaFold programme, it can be done in just two hours. After that, researchers can begin examining the suitability of drug molecules. Again, part of the work can be shortened and carried out through computer modelling programmes.”
The improved version of the AlphaFold programme, AlphaFold2, created and refined by the Nobel laureates from Google DeepMind, is freely available to everyone. By October this year, shortly before the prize announcement, it had already been used by over two million people across nearly 200 countries.
Photo: LIOS | Raitis Bobrovs, head of the Structural Biology and Drug Design Laboratory at LIOS
